Discovery & Development: Metabolomics
High-throughput targeted metabolomics: the next generation
With its potential to transform various industries and fields of research − including pharmaceuticals, preventive healthcare, agriculture and food safety − metabolomics is a rapidly growing field that has the potential to revolutionise our understanding of biological systems and disease mechanisms
Weiqiao Rao at BGI Genomics
Metabolomics is a field of study that involves the large-scale analysis of small molecules − known as metabolites − within cells, biofluids, tissues and organisms. The collective set of metabolites and their interactions within a biological system is referred to as the metabolome. Metabolomics is similar to genomics – the study of DNA and genetic information within a cell, and transcriptomics – the study of RNA and differences in mRNA expression. However, metabolomics focuses on the substrates and products of metabolism, which are influenced by genetic and environmental factors.
Mass spectrometry: the leading platform in metabolomics
Metabolomics tests are typically conducted using mass spectrometry (MS) or nuclear magnetic resonance (NMR). NMR can measure metabolite levels in intact tissue, making it a valuable tool for studying metabolic processes in vivo. However, NMR has limitations regarding sensitivity and the number of metabolites that can be detected. MS-based metabolomics is a highly sensitive and selective technique that can detect a wide range of metabolites in a single analysis. The data generated by MS-based metabolomics can be used to identify metabolites, quantify their concentrations and compare metabolite profiles between different samples.
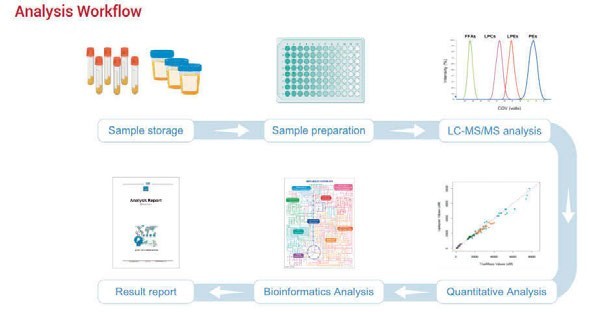
Figure 1: Workflow of high-throughput targeted metabolomics panel
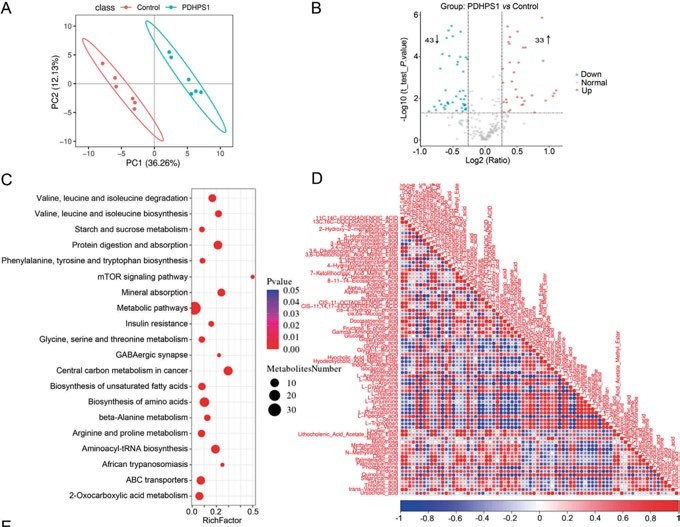
Figure 2a: Targeted metabolomics revealed that PDHPS1 reprogrammed metabolism in hypertrophic scar fibroblasts
Untargeted and targeted approaches in metabolomics
Targeted metabolomics is a more focused approach that involves identifying the chemical attributes of the metabolites to be tested before data acquisition occurs. Analytical methods are then designed to provide high precision, selectivity and reliability for detecting and identifying specific metabolites. Once the chemical identity of the metabolites is known, the subsequent process of deriving biological information from the acquired data can be started immediately after a complete overview. This knowledge can be used to design analytical methods specific to the metabolites of interest, enabling accurate detection and quantification.
The gap between untargeted and targeted metabolomics
High-throughput targeted metabolomics is a technique that allows for the rapid and efficient analysis of a large number of metabolites in a sample. It serves as a bridge between untargeted and targeted metabolomics by combining the advantages of both approaches. Untargeted metabolomics is a comprehensive analysis of all metabolites in a sample, but it can be time-consuming and may miss essential metabolites. Targeted metabolomics, on the other hand, focuses on a specific set of metabolites, but it may not capture the full metabolic profile of a sample. High-throughput targeted metabolomics allows for the analysis of a large number of targeted metabolites in an example while also providing information on other metabolites present in the sample. This approach can help to bridge the gap between untargeted and targeted metabolomics, providing a more comprehensive understanding of the metabolic profile of a sample.
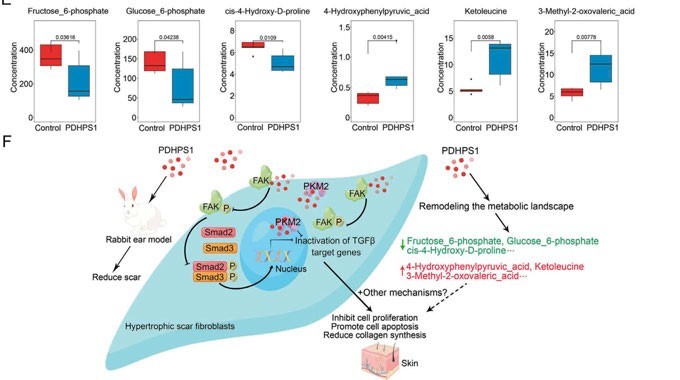
Figure 2b: Targeted metabolomics revealed that PDHPS1 reprogrammed metabolism in hypertrophic scar fibroblasts
The human metabolomics detection project: simultaneous detection and quantification of over 2,000 metabolites
Amino acids, bile acids and organic acids are vital substances with significant roles in human health − disturbances in their metabolism can lead to metabolic disorders and diseases. Identifying and quantifying these metabolites accurately is crucial, as they can provide valuable insights into biological processes, disease mechanisms and may also serve as potential biomarkers for diagnosis or treatment monitoring. However, identifying and quantifying important metabolites in metabolomics analysis can be challenging due to their low abundance in bio-samples. However, these metabolites may be present in very low concentrations in bio-samples, making them difficult to detect and quantify using normal analytical workflow.
As a result, specialised techniques such as enrichment, derivatisation or targeted analysis may be required to improve the detection and quantification of essential metabolites in bio-samples.
The Human Metabolomics Detection Project (HMDP) intended to identify and quantify more than 2,300 metabolites by derivatisation in a variety of biological samples, including plasma, urine and tissue. The targeted metabolites were designed from various metabolic pathways, which would benefit, for example, studying disease mechanisms. The combination of liquid chromatography and mass spectrometry (LC-MS) with the simple-to-use kit results in an accurate and sensitive assessment of metabolites (Figure 1).
Several known amounts of chemical compounds, ‘the internal standards’, must be added to each sample being measured in order to assure accuracy and reliability in metabolite quantification. Such as inconsistencies in sample preparation, instrument performance and other elements that may impact metabolite measurement can be corrected in this way.
The standard curve is another tool to ensure accurate quantification. It is created by examining a number of samples with known quantities of the target metabolite. A regression line can be produced by graphing the signal strength of each sample against its known concentration. The concentration of the metabolite in an unknown sample can then be determined based on the signal intensity of the metabolite by calculating the slope and intercept of the regression line.
A study that investigated how an endogenous peptide (PDHPS1) unregulated in hypertrophic scar-1 inhibits cell growth has used this approach.11The rabbit model was used to establish the hypertrophic scar fibroblasts and it was discovered that PDHPS1 strongly suppresses the formation of scars. 29 genes had changed expression patterns with RNA-seq and the proteome experiment discovered 46 proteins with higher expression. The main glycolysis intermediates (glucose-6-phosphate and fructose-6-phosphate) were discovered to significantly decrease, according to the results of the high-throughput targeted metabolomics analysis. Concluding the multi-omics results, the mechanism of PDHPS1 interaction with proteins, and the resulting remodelling of the metabolic landscape were reported (Figures 2a, 2b).
Overall, a comprehensive metabolomics solution is demanded for research applications, including biomarker discovery, drug development and clinical studies. The panel of metabolites and wide concentration range make it a powerful tool for identifying and quantifying metabolites in complex biological samples. With high sensitivity and accuracy, the high-throughput targeted metabolomics panel can provide valuable insights into metabolic pathways, disease mechanisms and has the potential to facilitate the development of new diagnostic and therapeutic approaches.
References
- Li Jet al (2023) Peptide deregulated in hypertrophic scar-1 alleviates hypertrophic scar fibrosis by targeting focal adhesion kinase and pyruvate kinase M2 and remodeling the metabolic landscape, Int JBiol Macromol 30.235.123809

Dr Weiqiao Rao is the director of R&D at the BGI Genomics, Mass Spectrometry Centre. He earned his PhD from the Technical University of Denmark, Denmark. Dr Rao has over ten years of expertise in developing and applying proteome and metabolome experimental methods and is dedicated to the translational application of mass spectrometry in medical research and clinical practice. Dr Rao has many SCI articles to his credit, as well as 13 patent applications and participation in a number of scientific research programmes, including the National Science and Technology Support Programme.