Genomics
Breaking Away From DNA Breaks
Base and prime editing’s applications in cell and gene therapy are continually increasing as scientists innovate the technology across the industry
Dr Andrea Frapporti at Horizon Discovery, a PerkinElmer company
Thanks to the groundbreaking technological advancements of the last decade, a range of gene editing agents are now at scientists’ disposal. The discovery of CRISPRs lies at the heart of this revolution, paving the way for faster, easier, and more precise manipulation of genomic sequences. While the vastness of their contribution to the field of genetic engineering is hard to overstate, some flaws limit their potency in therapeutic applications. Due to biological constraints, the controlled introduction of pinpoint, predefined single nucleotide variants (SNVs) or small insertions-deletions (indels) at target genomic locations remains highly inefficient in most mammalian cell types. This is particularly the case in post-mitotic cells, which are an important therapeutic target. Additionally, CRISPRs rely on the introduction of DNA doublestranded breaks (DSBs) via their associated nuclease – most famously Cas9 – to function. While tolerable in some research applications, this DNA lesion results in high cytotoxicity in many cell types, as well as an increased risk for potentially oncogenic aberrant chromosomal rearrangements.
To circumvent these issues, two pioneering next-generation gene editing agents have recently been developed: base editing and prime editing.
Base Editing
Between 2016 and 2017, the groups of Akihiko Kondo and David R Liu devised the first generation of CRISPR-Casmediated base editing systems to overcome the limitations of the standard CRISPR-Cas9 platform (1-2). Base editing consists of a deaminase enzyme fused to a Cas9 nickase (nCas9) to guide the introduction of SNVs via chemical modification, rather than physical replacement, of the target nucleotide in DNA. Two main classes of base editors have been developed: cytosine base editors (CBEs) and adenine base editors (ABEs). CBEs mediate the deamination of a cytosine into uracil, which has the base pairing properties of thymine. Following DNA replication, this results in a C-G to T-A base-pair conversion. ABEs, on the other hand, induce the deamination of an adenosine into inosine, which is read by the cellular DNA replication and transcription machineries as guanine, leading to an A-T to G-C substitution.
Since these SNVs are introduced without the need for hazardous DNA DSBs, this technology allows efficient editing in both dividing and terminally differentiated cell types, therefore expanding its versatility for research and therapeutic applications. A recent study estimated that around 37% of all pathogenic and likely pathogenic SNVs could potentially be corrected with current base editing architectures, accounting for approximately 4,000 genetic disorders (3). It is no surprise that cell and gene therapy research is showing great interest in base editing, from cancer immunotherapy through to the treatment of rare, debilitating genetic diseases.
In the ex vivo cell therapy space, base editing is a particularly promising approach for the generation of allogeneic CAR T therapies, where T cells from healthy donors are engineered to destroy cancer cells in patients. For allogeneic cell therapies to work, multiple genes must be edited to overcome the risk of graft-versus-host disease and for host-versus-graft disease. Multiplex base editing has been used to generate allogeneic CAR T cells in a number of studies, paving the way for new routes in cancer immunotherapy (4).
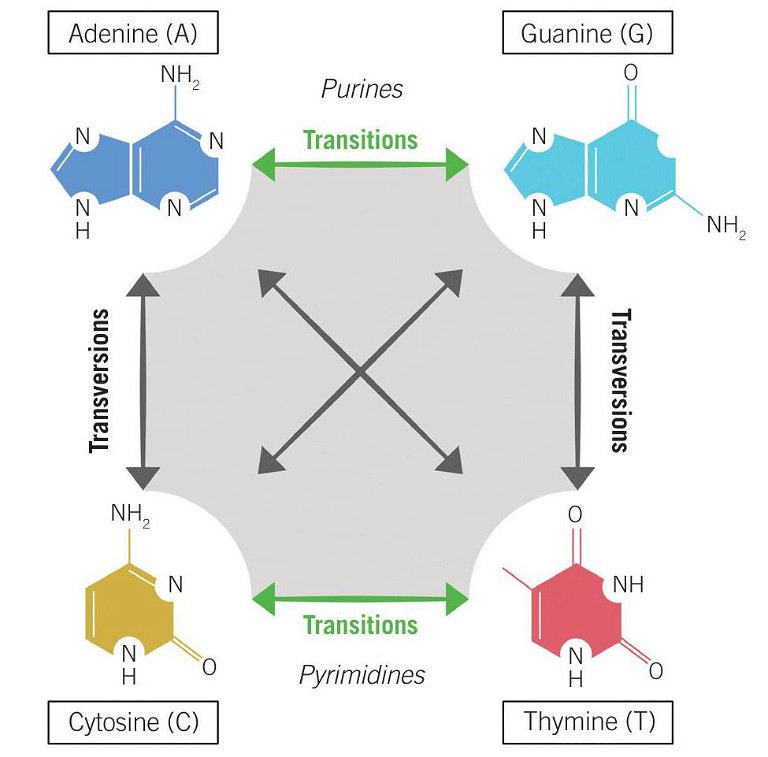
Figure 1: Transition vs transversion mutations
Base editing has also shown promise in reverting pathogenic SNVs and alleviating symptoms of monogenic disorders in cell lines, mouse, and non-human primate models. Preclinical research applications include sickle cell disease, hereditary tyrosinemia type 1, lysosomal storage disease, Duchenne muscular dystrophy, hypercholesterolemia, and Hutchinson-Gilford progeria, demonstrating the potential of base editing in therapeutic applications and offering hope to patients afflicted by these debilitating disorders (5-10).
Although base editing is currently limited to the four transition mutations (C→T, A →G, G→A, and T→C) (see Figure 1), rapid advances in technological development are widening the scope of its potential. A recent study began to expand the capability of base editing to include two transversions (C→G and G→C) (11). However, in order to fully expand the editing repertoire to include transversions and, crucially, small insertions and deletions which cannot be facilitated by base editing, a complementary tool is needed – enter prime editing.
Prime Editing
To address these unmet needs, David R Liu’s group at Harvard University developed an additional gene editing platform. Instead of the deaminase enzyme fused to nCas9 used in base editing, Anzalone et al employed a reverse transcriptase. A modified prime editing guide RNA (pegRNA) is provided to prime the reverse transcriptase to introduce the desired edit at the target location. The new system, aptly named prime editing, facilitates the introduction of predefined indels, once again without the need for DNA DSBs (11).
Ultimately, prime editing enables controlled and precise insertions of up to 44bp, deletions of up to 80bp, and the introduction of all 12 single base conversions, including the eight transversions that the base editing system currently falls short on (C→A, C→G, G→C, G→T, A→C, A →T, T →A, and T→G) (see Figure 1). Given this potentially wider editing scope, the implication is that prime editing could address ~90% of pathogenic variants in the ClinVar database, thereby increasing the range of targetable diseases without the need for generating DSBs (11).
Proof of concept studies applying prime editing to gene therapy did not lie in wait. A recent paper reported the use of prime editing to correct a small deletion responsible for Duchenne muscular dystrophy, which restored functional dystrophin expression in cultured human induced pluripotent stem cell-derived cardiomyocytes (8). Several other studies have already employed prime editing, either in vivo or ex vivo in cultured human cells, organoids, or mouse models for the correction of hereditary diseases including hereditary tyrosinemia type 1, metabolic liver disease phenylketonuria, or Wilson’s disease, demonstrating the high interest preclinical research has for this promising new tool for the treatment of afflicting genetic diseases (12-14).
Since the inception of prime editing in 2019, its applications in cell and gene therapy continue to grow as scientists explore new ways to adapt the technology to the field – aproof of how quickly this new tool has been adopted and how much interest it is raising in the genome editing space.
Comparison, Limitations, and Future Developments
Base and prime editing burst into the genetic engineering arena in succession, immediately providing a promising safer alternative to previous DNA DSB-inducing technology. Evidence produced so far points to a high degree of complementarity between the strengths and weaknesses of the two platforms. For instance, while base editors currently provide higher efficiency for the installation of precise SNVs, prime editing generally features lower bystander editing (see Bystander Editing boxout). Given that the highly modular architecture of prime editors requires a considerable locusspecific optimisation, base editing remains the favoured method for scenarios where bystander effects can either be avoided or are tolerated (15-16). When this is not possible, or when the desired edit cannot be generated via base editing (indels or transversion mutations), prime editing provides this important versatility.
Bystander Editing
‘Bystander editing’ occurs when additional cytosines or adenines in the immediate vicinity of the target base also become edited. While this can be tolerated when the desired outcome is the disruption of a gene, many therapeutic applications require very low, if not absent, bystander editing. The generation of less processive deaminases or the reduction of the activity window are some of the strategies undertaken to try and reduce this effect.
Overall, efforts to improve accuracy, reduce off-target and bystander editing, and expand the range of target genomic loci have been relentless since both technologies were created. Extensive optimisation has been undertaken to develop more accurate Cas9 variants and more specific and better performing deaminases, or more efficient pegRNA designs.
The delivery method for the use of base or prime editors as therapeutics is another crucial determinant of their efficiency and safety profile in vivo or ex vivo. For example, gene editing agents can be delivered as double-stranded DNA, mRNA, or ribonucleoprotein (RNP) complexes. Compared to a DNA-based delivery, mRNA or RNP delivery results in a quicker response and a more transient expression, thereby attenuating unintended off-target activity towards more therapeutically relevant levels. The choice of delivery vehicle is also pivotal for the safety and success of genome editing, especially when it comes to gene therapy applications. Adeno-associated virus (AAV) delivery is particularly popular in the field due to the low immunogenicity and toxicity, with two AAV-based gene therapies currently approved by the FDA. Because multiple serotypes have been identified having different tissue tropism, the use of AAVs also allows for a certain degree of tissue specificity.
One main obstacle in using this viral-based delivery for base and prime editing lies in their limited packaging capacity, making it impossible to fit the entire set of editors, RNAs, and relevant promoters into a single vector. However, scientists have developed innovative dual AAV systems to circumvent this issue (7-8,10-11,13). These are based on the use of split inteins, which function as protein introns allowing the splicing and subsequent fusion of two separate peptides into a full-length protein. Split editors are delivered as two separate fragments, which are then fused intracellularly, thereby reducing the packaging size of individual AAV vehicles. Future studies will need to establish safety profiles and demonstrate editing efficacy at different viral doses for large animal models.
On the Horizon
The recent addition of base and prime editing to the gene editing toolbox has opened new, exciting, and promising paths for the development of safer, DSB-independent genome editors. While still in their infancy, these highly complementary technologies hold great promise for the progress of gene and cell therapies and for fundamental research into in vitro disease modelling. Their relevance for the treatment of most human genetic disorders, caused either by SNVs or small indels, is undeniable. However, with great power comes great responsibility. Future studies will need to assess base and prime editing safety profiles genome-wide, improve their efficacy at different genomic sites in different models and validate the clinically relevant delivery methods. With research off to an exciting and promising start, both gene editing technologies now lie under the spotlight. The outcomes of future advancements will determine how they will be employed in gene and cell therapy applications.
References
- Komor AC et al, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature 533(7603): pp420-4, 2016
- Nishida K et al, Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems, Science 353(6305): aaf8729, 2020
- Lavrov AV et al, Genome scale analysis of pathogenic variants targetable for single base editing, BMC Med Genomics 13(8): p80, 2020
- Harbottle JA, Immunotherapy to get on point with base editing, Drug Discov Today S1359-6446(21)00190-2, 2021
- Newby GA et al, Base editing of haematopoietic stem cells rescues sickle cell disease in mice, Nature 595(7866): pp295-302, 2021
- Rossidis AC et al, In utero CRISPR-mediated therapeutic editing of metabolic genes, Nat Med 24(10): pp1,513-8, 2018
- Bose SK et al, In utero adenine base editing corrects multi-organ pathology in a lethal lysosomal storage disease, Nat Commun 12(1): p4,291, 2021
- Chemello F et al, Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing, Sci Adv 7(18):eabg4910, 2021
- Musunuru K et al, In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates, Nature 593(7859): pp429-34, 2021
- Koblan LW et al, In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice, Nature 589(7843): pp608-14, 2021
- Anzalone AV et al, Search-and-replace genome editing without double-strand breaks or donor DNA, Nature 576(7785): pp149-57, 2019
- Kim Y et al, Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease, Cell Stem Cell S1934-5909(21): 00167-3, 2021
- Böck D et al, Treatment of a metabolic liver disease by in vivo prime editing in mice, bioRxiv p. 2021.08.17.456632
- Schene IF et al, Prime editing for functional repair in patient-derived disease models, Nat Commun 11: 5352, 2020
- Scholefield J and Harrison PT, Prime editing – an update on the field, Gene Ther 28(7-8): pp396-401, 2021
- Geurts MH et al, Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids, Life Sci Alliance 4(10): e202000940, 2021

Dr Andrea Frapporti is a scientist in the Base Editing Department at Horizon Discovery, a PerkinElmer company, where he works as part of the R&D team focused on the development and commercialisation of a novel base editing platform. Prior to joining Horizon Discovery, Andrea worked at the Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge, and gained his PhD in genetics at the University of Paris Diderot, France.