Discovery and development: gene synthesis
Next-generation size selection: accelerating the pace of gene synthesis
Synthetic biology has been a cornerstone in recent health advancements, including drug discovery, development and manufacturing, however there are challenges facing this market
Jennifer Golding at Yourgene Health
Synthetic biology has cemented itself as a cornerstone in global health advancements. By combining our understanding of nature with technological innovations and genetic engineering principles, it plays an increasingly pivotal role in accelerating drug discovery, development and manufacturing processes as well as other research disciplines. However, maximising the purity of lengthy constructs is the biggest hurdle to gene synthesis, in a market that increasingly demands complex, quality DNA on tight turnaround times.
The chemical synthesis of DNA base by base is an immensely powerful tool, especially when no template is needed. Being able to build naturally occurring sequences and variants, as well as de novo constructs that do not exist in nature, offers almost limitless possibilities. The subsequent assembly of these synthetic sequences into genes, with the ability to create any number of systematic or random mutants more efficiently and with greater accuracy than with traditional mutagenesis, has become a key enabling technology across multiple market sectors.
It is easy to see how improvements in gene synthesis methods could greatly accelerate the classic design-build-test-learn-repeat cycle (Figure 1) and enable the next-generation of therapeutics to reach patients sooner.
Rapid pharmaceutical response
Using gene synthesis processes, parts of (or, indeed, entire) pathogenic genomes can be synthesised for deployment in multiple synergistic tactics. Full-length attenuated genomes help develop vaccines against viral and bacterial infections (eg, yellow fever, influenza and oral polio, tuberculosis, BCG and oral typhoid). While annual threats like influenza require novel, strain-specific antigens to ensure the vaccine formula is effective against seasonal variants.
The events of both the original COVID-19 pandemic and the subsequent waves of new variants highlighted, more starkly than ever, that epidemiological events necessitate rapid pharmaceutical response, at scale, and the role that gene synthesis can play in this. In scenarios where multiple variants emerge in tandem, gene synthesis can help scientists to rapidly generate strain-specific antigens (eg, spike proteins) to support the development of vaccines and therapeutic antibodies to treat patients. It is clear that optimising the biomanufacturing processes to deliver effective prophylactics and therapies in a timely manner is critical in controlling both annual or localised outbreaks and emerging variants before they become epidemics.
How is gene synthesis performed?
Gene synthesis involving the assembly of designed oligonucleotides requires the following steps (Figure 2):
1. DNA sequence selection: select the gene of interest and design the sequence to be synthesised. Very large synthetic genes will typically be divided into 500–1000 kb chunks and assembled later
2. Sequence optimisation and oligonucleotide design: sequence analysis is required to determine the best way to divide the gene or chunks into oligonucleotides fragments, or ‘oligos’, for synthesis
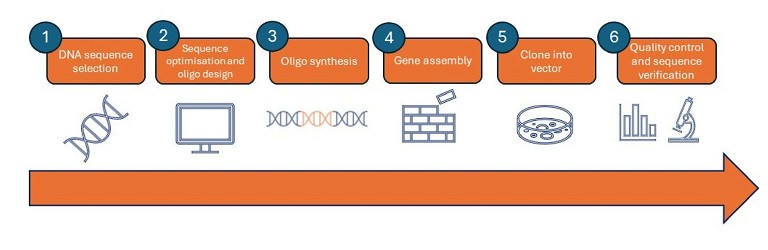
Figure 1: The design-build-test-learn–repeat cycle
3. Oligo synthesis: the stepwise addition of nucleotide monomers to form short oligos using modified nucleotides, called phosphoramidites. This method ensures that nucleotides assemble in the correct way and prevents the growing strand from engaging in undesired reactions during synthesis
4. Gene assembly: polymerase chain assembly is a standard technique for polymerase-based oligo assembly in a thermocycler (sometimes known as templateless PCR). The principle is to combine all of the single-stranded oligos into a single tube, perform thermocycling to facilitate repeated rounds of annealing, extension and denaturation, and then use the outermost primers to amplify full-length sequences
5. Clone into vector: synthetic constructs are then cloned into a vector and inserted into a bacterial host, such as Escherichia coli (E. coli), to amplify the sequence for analysis
6. Quality control and sequence verification: due to the inherent potential for error in each step of the gene synthesis process, all synthetic constructs should be verified before use. Sequences harbouring mutations must be identified and removed from the pool or corrected. Internal insertions and deletions, as well as premature termination, are common in synthetic DNA sequences.
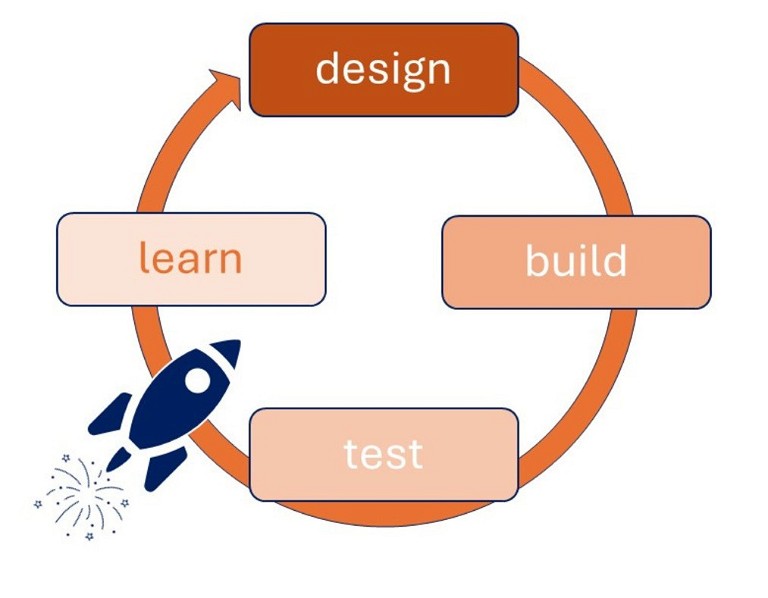
Figure 2: Gene synthesis overview
Challenges of gene synthesis: long fragments
Many of the high-value target sequences for which gene synthesis is employed are >3kb in length. This level of complexity greatly complicates synthesis and necessitates a hierarchical, iterative build (Figure 3).
Commonly, these synthetic oligos are joined to assemble the full-length genome as a bacterial artificial chromosome (BAC), with organisms like E. coli acting as the bacterial host to make the synthetic products ready to use in various applications. Placing synthetic genes into a vector:
• Protects the DNA against degradation during storage
• Allows complete sequence verification using primer sites on the vector that flank the gene insert
• Facilitates clonal amplification in a transformation-competent bacterial host
• Facilitates shuttling of the synthetic gene into expression vectors for transfection or electroporation into host cells of interest for transient or stable expression.
However, to get to this point, researchers must employ a process using bench techniques that have been around for decades. The process is predicated on synthesising a construct that is as pure as possible, and then achieving the desired quantity by cloning it. This, in itself, requires time to let colonies grow after being plated out, before finally beginning the laborious task of isolating the exact construct of interest from the colonies. There is also inherent potential for the introduction of impurities and several steps further down the line, which jeopardise the chances of isolating your construct, not least when transforming it into a bacterial host.
Despite improvements over the past several decades in recombinant DNA tools, such as enzymes and cloning vectors, even the seemingly simple task of isolating a gene using PCR cloning or restriction digestion can be tedious and error-prone, all the more so with sequences of great length and/or complexity. While synthesis by certain chemistries does have very low error rates, errors do accumulate as strand length increases; therefore, gene synthesis typically uses oligos with lengths of 40–200bp. Optimal oligo length depends upon the assembly method that will be used, the complexity of the sequence and the operator’s preferences.
Jumping the hurdle: size selection
When the fidelity of the constructs produced from the desired sequence of interest cannot be guaranteed, the difficulty in isolating the construct at the end of the process is many orders of magnitude more difficult and time-consuming. It is this issue of purity that is the largest hurdle to manufacturing complex, quality DNA at increasingly tight turnaround times demanded by industry.
One way of minimising the extent to which time is spent verifying the correctness of the synthetic sequences produced is to implement a process known as size selection. This process is about enriching and purifying; preferentially selecting material of interest by differentiating based on size. Researchers can use size selection to minimise noise associated with concatenated truncation products, which are an unavoidable consequence of inefficiencies in both oligo synthesis and PCR assembly. Given that off-targets get produced much more often than desired products, taking a sample from construct to clone is often not of adequate purity because the processes used to synthesise it are imperfect. The accumulation of errors from phosphoramidite chemical synthesis alone can mean only ~30% of any synthesised 100-mer is the desired sequence.
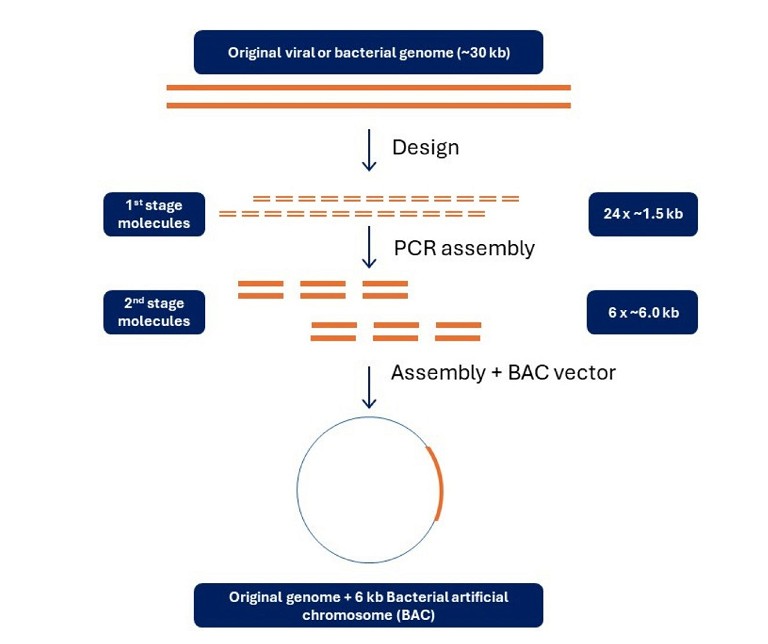
Figure 3: Synthetic sequence design and assembly
Prematurely terminated strands can be removed by purifying the eluted product via gel electrophoresis and cutting out the band with the correct length following oligo synthesis (Figure 2: Step 3). Automated gel electrophoresis approaches that allow for selection of desired fragments from 20bp–10kb, can deliver high yields with precision and at scale.1
Additionally, the presence of numerous concomitant truncation products frequently results in a success rate below 10% in complex synthetic reactions, meaning that <10% of the bacterial colonies from which the construct is harvested actually contain the desired insert. Since this cannot be known until after sequencing, this is a huge time sink and headache to operators. Size selection at the end of the gene assembly process (Figure 2: Step 4) can increase that to 90%. By doing this, checking and sequencing of fewer colonies is required before the true positive construct is found. When the efficiency of one of the most laborious stages of a very time-consuming process is greatly enhanced, the turnaround time is decreased dramatically. When longer DNA constructs are needed, size selection technology can be used to maximum effect. Tools that enable researchers to scale operations by allowing multiple size selections to be performed at the same time can help to truly revolutionise the traditional approach to gene synthesis.
Get to the DNA you want
Exploiting size selection in short fragment applications has already solidified it as an indispensable technique in clinical settings. It first demonstrated its efficacy in optimising sensitivity of results gained from non-invasive prenatal testing (NIPT) and oncology investigations, where fragments of <150bp are pivotal and have the added bonus of being available from liquid biopsy (peripheral blood draw) rather than more invasive sample types.
Now, with the increasing popularity of long-read approaches to whole genome sequencing and its adoption within clinical settings, providers such as PacBio and Oxford Nanopore Technology are also realising the value of size selection in both research and clinical settings for the isolation of fragments 3-30 kb and beyond.2 Generating larger constructs has greater economic value for pharmaceutical companies who need to make vaccines and therapies faster than ever before to meet increasing demand. The gene synthesis industry as a whole is trending towards turbocharging these enhanced biomanufacturing processes, with size selection a key asset in doing so.
Where speed, complexity and cost matters, next-generation size selection technologies that deliver the highest degree of automation alongside scalable, precise and robust electrophoretic analysis offer clinical and research groups a viable option for the analysis of DNA constructs at high volumes.
References
- Nesbitt M (2023), ‘A Ranger TechnologyApplication: Synthetic Biology and Gene Synthesis’, Your Expert
- PacBio (2023),Size Selection ofPacBio SMRTbellLibraries withthe LightBenchInstrument, Technical Note

Jennifer Golding is a product manager at Yourgene Health, an integrated technologies and services business, enabling the delivery of genomic medicine. Jen has eight years’ NHS experience in Cellular Pathology, having attained the Certificate of Competence in Cervical Cytology in 2011 and pursued her special interests in HPV testing and male fertility analysis. Now with four years’ commercial experience as a product manager in the molecular diagnostics industry, Jen has worked with medical devices including NGS, FISH probes, PCR assays and NIPT technology. Jen has a BSc in Medical Biochemistry from the University of Birmingham, UK and is an HCPC registered biomedical scientist.