Discovery & Development: Cell Identity
Coding of Cell Identity
Non-human cells have been used in drug discovery for years. How are novel technologies helping researchers to move past this method?
Emmanouil Metzakopian at bit.bio
What gives a cell its identity? This fundamental research question has shaped my career for 17 years. In seeking answers, science is paving the way for new generations of cell-based therapies to alleviate our burden of disease. Since my earliest days in the laboratory, my focus has been getting therapies to patients. The advice from my mentors was, first and foremost, to truly understand cells:
• How are they born?
• How do they grow?
• How do they get their identity?
I started studying the mouse midbrain, where dopaminergic neurons are formed. The relevance of dopaminergic neurons in Parkinson’s disease drove me to dig more deeply into developmental questions around dopaminergic neuron specification and differentiation. Parkinson’s disease is a consequence of dopaminergic neuron loss; I therefore challenged myself to learn how to create new ones in vitro.
Many diseases are caused by gene mutations. A mutant gene will change the function of a protein, which will have a direct molecular consequence, and the first arena where this consequence will play out is on the cellular level. By manipulating cells’ genetic mechanisms in our explorations both of basic science and of medical research, we are recognising the advantages of cells over small-molecule drugs, ushering in regenerative approaches and a future in which ‘smart’ cells can be programmed to recognise and attack disease. In fact, this is already happening with so-called ‘CAR T therapies’ – human T-cells reprogrammed to seek out and destroy cancers.
By modelling gene mutations in cells, we can try to recapitulate the disease process, understand it and explore ways of treating it. But of course, the cell identity and physiological relevance is critical for modelling any given disease.
Take for example the neuroligin-4 gene, which codes for a synaptic adhesion molecule NLGN4. This gene has long been of interest to scientists studying familial autism spectrum disorder (ASD), with a nearly 100% penetrant
ASD phenotype arising from over 50 distinct mutations in NLGN4. However, no NLGN4 homolog is found in Rattus norvegicus, so how to study it? In the mouse, a poorly conserved ortholog, ‘NLGN-like’, is involved in inhibitory synapses in the spinal cord and brainstem. In humans, the gene is primarily expressed in excitatory forebrain synapses.1
This contrasting finding in neuroligin-4 provides just one example of poor evolutionary genetic conservation and low translatability of animal research to human health and disease. A more classic manifestation is the high failure rate of drugs developed to treat Alzheimer’s disease – drugs initially tested in mice, despite mice lacking the ability to develop the disease. For impactful human medical research, what we need are human-relevant cell models that recapitulate human disease. The outdated and inappropriate use of animal models is now recognised in the 2022 FDA Modernization Act 2.0, which welcomes human-translatable in vitro models earlier during drug discovery and eliminates the federal mandate for animal testing. With in vitro models now approaching the necessary reproducibility and disease-context fidelity, the likelihood of successful human drug discovery is rising.
For clarifying our basic understanding of what makes cells tick, some useful workhorses are the classic HeLa, HEK293 and some immortalised T cell lines, which have the advantage of being easy to grow in vast quantities. However, when we move beyond cellular biology into medical research, a different playground is required. Human-induced pluripotent stem cells (hiPSCs) provide the platform for today’s most human-relevant research. At this point, scientists start to care about cell identity: the cell function, context and any disease-relevant mutations. By modelling disease in cells with exactly the right identity, genetic screening or expression profiling to identify therapeutic targets acquires a greater level of precision.
Despite offering great promise, protocols for differentiating hiPSCs to a target cell fate still bring some bottlenecks to the bench. Directed differentiation protocols, in which stem cells are shepherded through a series of stages to become a new cell type of interest, are very labour-intensive. These protocols essentially try to mimic embryonic development in a dish via the carefully scheduled application of patterning factors, growth factors and small molecules. These activate or inhibit certain signalling pathways to give rise to authentic populations of the target cell type. Because of the complexity of these protocols, their user-to-user reproducibility is low. This is because many factors can affect the outcome, such as the initial cell density, how the cells are dispersed and how they are counted. As a result, the final cell population can be highly variable and heterogeneous, with differences in efficiency and purity.
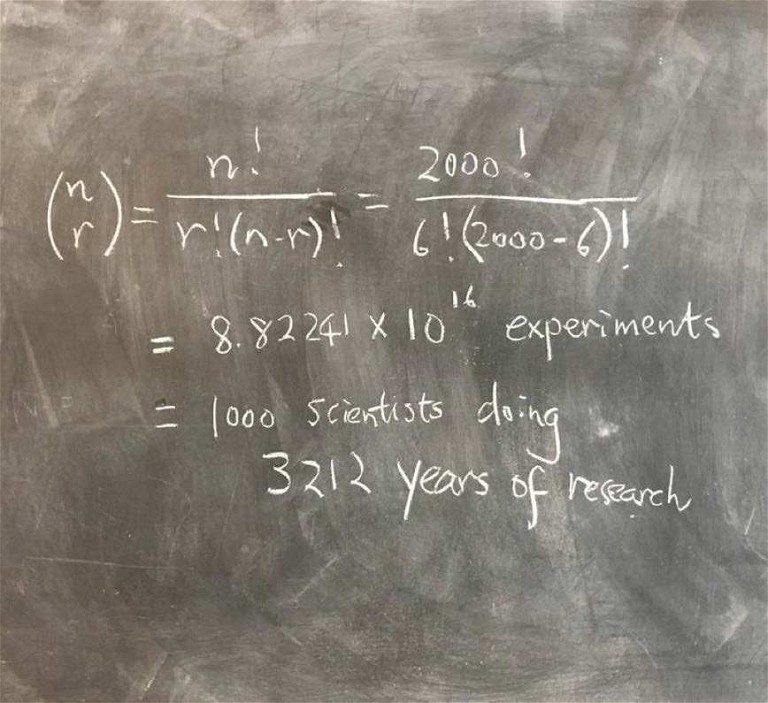
Figure 1: Identifying transcription factor codes for cell identity is no easy feat
In drug discovery pipelines, human iPSC-derived cells offer much needed physiological relevance, however these models can be hindered by low scalability, heterogeneous populations and long complex protocols, all compromising data quality. Investigators who witness this problem on an industrial scale in companies performing drug discovery are constantly searching for optimised protocols that offer greater consistency and lot-to-lot reliability.
Dr Marijn Vlaming, Head of Biology at Charles River Laboratories, says, “One of the most important challenges is the standardisation or reproducibility of the cell model. That is something the whole community has been struggling with when using iPSC-derived models.”
Production time is a factor too. “It’s a very time-consuming activity to develop cell models from iPSCs. During drug discovery, when you need a lot of cells because you want to test a lot of different compounds at a high throughput, time is a really important parameter.”
“Whole-genome high-throughput genetic screens can be revolutionary: this process produces millions of cells with exceptional quality, consistency and purity”
Luckily, new technologies such as ‘precision cellular reprogramming’ are emerging that offer a solution to current iPSC-derived cell generation methods. Precision cellular reprogramming technology combines an inducible gene expression system with a unique combination of transcription factors, which when expressed in iPSCs drives the consistent adoption of a new cell identity. It is powered by intrinsic signals from transcription factors, overcoming other cell-generation methods’ inherent limitations caused by interference from external environmental factors, and consequently resulting in cells that are consistently reprogrammed into a highly defined cell identity and population.
Whole-genome high-throughput genetic screens can be revolutionary: this process produces millions of cells with exceptional quality, consistency and purity. With the introduction of iPSC-derived cells via reprogramming technology, the entire process takes just a matter of weeks, ultimately providing us with access to hundreds of millions of cells that meet exact specifications. Therefore, higher quality data can be delivered to clients.
New technology is an inducible gene expression system, so we can amplify iPSCs to billions of cells, then switch it on and watch as every cell consistently reprogrammes into the cell type dictated by specific transcription factors.
With this highly synchronous and consistent precision cellular reprogramming technology, the required scale-up to billions of human cells for disease modelling and high-throughput drug screening applications, or even to trillions of cells for therapeutic purposes, can proceed with greater speed, consistency and physiological relevance. These advantages facilitate the translation from laboratory to clinic, and from clinic to patient. This game-changing technology fills me with excitement for cell therapies; for example, in Parkinson’s disease, transplantation of dopaminergic neurons offers patients hope for the future.
With consistent, disease-relevant cell models now entering our playground, and with their gradual scale-up and cost benefit delivering comparable availability to HeLa and other workhorses, biotech pioneers are focusing on diseases that have previously been difficult to mimic in vitro.
Three-dimensional co-cultures that recapitulate human neurodegenerative disorders could provide a much more promising environment for Alzheimer’s research than any animal model. This won’t just affect research and drug discovery: cell therapy will be revolutionised as a major bottleneck of access to cells consistently at scale is removed.
If cell identity is a product of certain transcription factors being switched on, how do we establish which transcription factors code for which cell type? Our next challenge, then, is to identify these unique combinations, of which there are 8.8 x 1016, assuming that my calculations are correct (Figure 1)! No easy feat. Some of the transcription factors that researchers have stumbled upon have proven to be very powerful lineage-determination factors. But being able to predict the precise code that defines cell fate would open up an entirely new level of capability to the technology.
References
1. Visit: https://pubmed.ncbi.nlm.nih.gov/31257103/

Dr Emmanouil Metzakopian is currently the vice president of Research and Development at bit.bio, a synthetic biology company providing human cells for research, drug discovery and cell therapy. Dr Metzakopian studied biochemistry and biotechnology at the University of Thessaly in Greece, before obtaining his PhD from University College London in 2010 and completing his postdoctoral training at the Wellcome Sanger Institute. Dr Metzakopian joined bit.bio in 2021 as the Head of Innovation and was appointed as vice president of Research and Development in 2022.