Analytics: Single-Cell Analysis
Droplet-based microfluidics advances for single-cell analysis
Droplet-based single-cell analysis systems provide an invaluable tool to accelerate research and development. Recent scientific advances enhance the applicability of these droplet-based systems to address the age-old problem of characterising, understanding and selecting cells
Richard Hammond at Sphere Fluidics
Droplet-based systems for single-cell analysis have become an established method for R&D − particularly for identifying, analysing and isolating specific target cells in large populations. Using microfluidics to encapsulate cells in droplets provides critical benefits in an R&D context compared with traditional bioprocessing methods. These benefits include: rapid analysis to enable large cell populations to be screened in a short space of time, localisation of secreted molecules to extend applicable assay types, a low-shear stress environment to protect cells and ultra-miniaturisation to drive down cost.
The droplet-based approach is being developed further to include new functions and techniques, such as new in-droplet single-cell characterisation and assessment modalities beyond fluorescence (such as mass spectroscopy and advanced imaging methods); functional assays examining and assessing the interactions between cell types; integration of gene editing methods into the droplet format; and the use of double-emulsion droplet systems to extend the technology to more hydrophobic molecules.
Characterising, understanding and selecting individual cells is fundamental to biological R&D. Since the invention of the first microscope in the sixteenth century, biological samples have been studied closely, as demonstrated through the work of Leeuwenhoek in the mid-to-late 17th Century. 1 Many technologies have since been applied to advance research, both to increase the amount of information obtained from each cell and to increase the speed at which cells can be characterised. However, analysing single cells in the 21st Century using traditional approaches requires multi-step operations and instruments, high hands-on time and lengthy liquid handling processes, as well as complex, low throughput bioassays for functional validation and verification.
One approach to these problems, introduced in the last decade, is the use of droplet-based systems, which builds upon ideas from flow cytometry for the rapid assessment and selection of individual cells. Cells are encapsulated in droplets of a suitable aqueous media contained in an oil carrier fluid and stabilised with a surfactant. The droplets are produced using microfluidic methods, where liquids are pumped through carefully designed geometric channels at the micrometre scale (Figure 1).
By varying the relative flow rates of the cell suspension and oil carrier fluid, the distribution of the number of cells per droplet can be controlled − particularly to obtain predominantly single cells per droplet based on the Poisson statistical distribution.
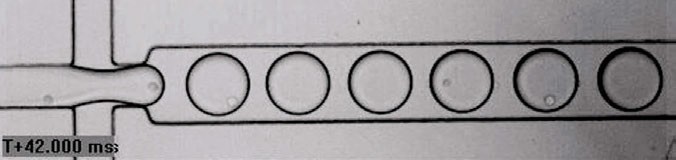
Figure 1: Encapsulation of single cells using droplet-based systems
These droplets can then be subjected to different operations all within the closed microfluidic system. Encapsulating cells in droplets brings many improvements to cell characterisation and selection methods. These include:
• Automation: The flow of droplets through microfluidic channels provides a robust and adaptable platform to automate complex workflows. Multiple processes such as encapsulation, incubation, splitting, combining and sorting can be integrated and carried out reliably and repeatably with minimal user input
• Speed and scale: Thousands of droplets can be created and processed per second. This allows large populations of cells to be interrogated in a few hours, enabling researchers to extend the scope of their work substantially
• Localisation: The individual droplets form a self-contained reaction vessel whereby the leakage rate of biomolecules is very small. Firstly, this allows the assessment of secreted molecules from cells, not just surface markers, which retain their physical location on the cell. Secondly, the small dimensions of the droplet (tens to hundreds of picolitres) allow a rapid increase in concentration of molecules at a given rate of expression, allowing strong signals to be developed quickly during assessment, thus increasing the detection sensitivity
• Benign environment: Cells are encapsulated in their preferred medium and the oil phase maintains the necessary gas exchange flows to allow respiration, thus the microenvironment can be precisely defined and controlled according to the cell type. Furthermore, the droplet shields the cell while it is being moved through the microfluidic channels at high speed, ensuring the cell is not exposed to detrimental shear stress despite the high velocity necessary to sustain overall throughput. These factors ensure cells can be assessed and selected while maintaining high long-term viability to support ongoing scale-up
• Cost: The small size of the droplets requires minimal amounts of reagents and buffers, meaning large-scale experiments can be performed cost-effectively as well as quickly. Similarly, the reduced need for user input and increased efficiency of the analysis process increases the overall cost-effectiveness and productivity, yielding longer-term economic benefits.
The classic droplet-based workflow for cell characterisation and selection integrates several individual process steps. A population of cells is encapsulated at, on average, one cell per droplet. The aqueous phase contains reagents to support a fluorescent assay, such as Förster Resonance Energy Transfer (FRET) or a binding assay with beads, which selectively identifies the expression of a molecule of interest (eg, an antibody) (Figure 2).This fluorescent signal is then used to separate low-expressing droplets from high-expressing ones by passing each individual droplet through detection optics and directing it into one of two channels depending on the signal. 2
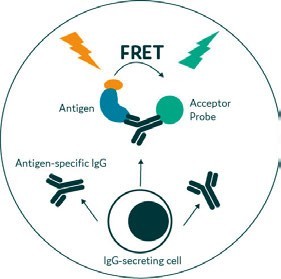
Figure 2: FRET-based detection of antibodies within single cell-encapsulated droplet
While this is currently the mainstay workflow used for droplet-based systems, there are several new techniques and approaches that offer the possibility of applying the technology – and realising the benefits – into new areas of pharmaceutical and biotechnology research.
Cell characterisation
Fluorescent methods require labelling which is not always suitable as this can disrupt the interaction of interest by changing the kinetics of the system, or the target molecules cannot be labelled. There are two interesting approaches that avoid this problem − the first of which is real-time brightfield imaging. Developments in camera and image processing technology now allow images to be captured and processed fast enough to support the assessment and selection of cells in droplets in real time.3 The droplet assessment rates of 4Hz (droplets per second) that are currently achievable are limited by the resolution and exposure time of the camera to capture still images for analysis. Given this, imaging methods are not a direct replacement for fluorescence at this juncture, but give an interesting secondary method, which is particularly suitable for removing the inevitable Poisson-derived empty droplets to enrich incoming cell populations.
The second is the integration of mass spectrometry methods; splitting droplets into two parts is easily done using microfluidics. By splitting a droplet, one part can be passed to a mass spectrometer and a direct assessment made of the molecular contents. The second part is retained in a flow channel and the mass spectrum signal used to decide if to retain or discard the second part for further analysis. This approach has been demonstrated for small molecules, peptide and proteins with assessment rates up to 30Hz.4 Mass spectroscopy thus has the potential to provide a non-fluorescent high-speed assessment method for droplets.
Implementing functional assays
Rather than isolating single cells and characterising the cell on its own, cells are co-encapsulated into droplets to bring two different cell types together thus more accurately simulating the in vivo conditions. This method opens up possibilities in immuno-oncology and neurobiology research where the interactions between cells are fundamental to understanding the disease-indicated mechanisms of action and for developing novel therapeutics. Examples include: assessing T-cell receptor mediated T-cell activation by co-encapsulating T cells with target cells; and characterising astrocyte responses to microglia-produced amphiregulin to identify potential therapeutic approaches for multiple sclerosis.5,6
Gene editing
Transfection techniques can be used to carry out edits on individual cells in their own microenvironment. By bringing the cell, plasmid and reagents into very close proximity in a tightly controlled environment, improved editing efficiency can be achieved at low cost.7 As well as implementing traditional techniques in a more effective way, droplets also allow more novel approaches, such as mechanoporation, whereby cells in droplets can be mechanically permeabilised to allow ingress of plasmid.8 Furthermore, droplet microfluidics methods naturally support library-based editing approaches using tools such as CRISPR. For example, cells can be exposed to a complete library of single-guide RNAs and specific edits that give the desired phenotypical response that can be selected for.9
The classic single-emulsion system (aqueous droplets in oil) is relatively easy to make using straightforward microfluidics and works well for hydrophilic molecules, as they remain in the aqueous phase and loss of analyte from the droplet is small. However, lipophilic or hydrophobic molecules are more challenging as they migrate away into the oil phase. By creating a double emulsion (typically aqueous droplet in oil droplet in aqueous carrier), the lipophilic molecules are retained in the thin oil layer. This opens up analysis of more complex biochemistry, for example investigation into antimicrobial peptides.10 The reverse system (oil in water in oil) has also been investigated as a method to extend the applicability of mass spectrometry methods.11
Droplet-based single-cell analysis systems provide an invaluable tool to accelerate research and development. Recent advances enhance the applicability of droplet-based systems to address the age-old problem of characterising, understanding and selecting cells. These advances extend the remit of droplet systems into areas such as immuno-oncology, neurobiology and synthetic biology, bringing the benefits of the approach to a wider and more diverse group of researchers in both the pharmaceutical and industrial biotechnology industries.
References
- Visit: makingscience.royalsociety.org/people/na8438
- Josephides D et al (2020) Cyto-Mine: An Integrated, Picodroplet System for High-Throughput Single-Cell Analysis, Sorting, Dispensing, and Monoclonality Assurance, SLAS Technoogy 25.2, 177-189
- Sesen M et al (2020) Image-Based Cell Sorting Automation in Droplet Microfluidics, Scientific Reports 10.1, 8736
- Kempa E E et al (2020) Coupling Droplet Microfluidics with Mass Spectrometry for Ultrahigh-Throughout Analysis of Cmplex Mixtures up to and above 30 Hz, Analytical Chemistry 92.18, 12605-12612
- Segaliny A I et al (2018) Functional TCR T cell screening using single-cell droplet microfluidics Lab on a chip 18.24, 3733-3749
- Wheeler M A et al (2023) Droplet-based forward genetic screening of astrocyte-microglia cross-talk, Science 379.6636, 1023-1030
- Pérez-Sosa C et al (2022) Single cell transfection of human-induced pluripotent stem cells using a droplet-based microfluidic system, Royal Society Open Science 9.1, 211510
- Joo B et al (2021) Highly effective transfection of human primary T Lymphocytes Using Droplet Enabled Mechanoporation, ACS Nano 15.8, 12888-12898
- Yu X et all (2023) CRISPRi-microfluidics screening enables genome-scale target identification for high titer protein production and secretion, Metabolic Engineering 75, 192-204
- Nutti N et al (2022) A Multiplexed Cell Free Assay to Screen for Antimicrobial Peptides in Double Emulsion Droplets, Angewandte Chemie (International ed. in English) 61.13, 202114632
- Heiligenthal L et al (2022) Analysis of double emulsion droplets with ESI mass spectrometry for monitoring lipase-catalysed ester hydrolysis at nanoliter scale, Analytical

Richard Hammond is chief technical officer at Sphere Fluidics, a company developing single-cell analysis systems underpinned by its patented picodroplet technology. Richard has over 20 years’ experience in managing the development of novel commercial products for healthcare and life sciences. He has held numerous senior positions, including leading product and technology development at Alere Inc, Cambridge Consultants Ltd and DNA Electronics Ltd. With extensive technical expertise, he has worked in cutting-edge technical areas, such as isothermal molecular diagnostics, automated cell transfection, CAR T-cell therapy manufacture and digital data storage in DNA. Richard holds MA and MEng degrees in engineering from King’s College, University of Cambridge, UK.