Discovery and Development: Computer Vision in Oligonucleotide Synthesis
From ACGT to AI: visionary advances in oligonucleotide synthesis
Considering the importance and cost associated with oligonucleotide-based therapeutics, how can innovative computing imaging techniques be used to aid in the process, while also creating an audit trail and increasing the data derived from these experiments?
Cei Provis-Evans at Reach Industries, and Sam Whitmarsh at CatSci
Oligonucleotide-based therapies are a major growth area, with the market estimated to grow from around $5bn in 2020 to $26bn by 2030.1 Oligonucleotides themselves are short lengths of synthetic DNA or RNA, either singleor double-stranded. They work in similar ways to our own native genetic material to modulate the activity of cells, for example by promoting or inhibiting the expression of a protein associated with a particular disease. One key advantage of oligonucleotide therapies over small molecule drugs is their high specificity. Their base sequence can be precisely designed to interact with the target gene, leaving other tissues unaffected. However, these therapies are costly and difficult to produce due to their complexity, and the expensive reagents and equipment required for manufacturing.
Considering the importance of the sequence to the efficacy of these therapies, it is vital that synthetic methods are equal to the task of ensuring an oligonucleotide contains the correct components bases. Equally important is the even more subtle challenge of ensuring that the correct bases are connected in the correct order. Considering this additional potential for failure, ensuring the quality and accuracy of synthesised oligonucleotides during drug development demands significant attention, rigour and expertise from the responsible scientists. This becomes even more critical as therapies progress to later stages of development. AI-enabled computer vision offers a significant opportunity to alleviate some of this burden: first by aiding in the synthesis process itself; second by providing an audit trail of the physical process, allowing for easy verification if uncertainties arise; and third by multiplying the quality and quantity of data derived from experiments.
To illustrate the potential value of computer vision in oligonucleotide synthesis, a generalised procedure for synthesising oligonucleotides will be shown, using the widely adopted solid-phase phosphoramidite method at a lab scale, which will highlight key areas where computer vision could transform the process.
Reducing errors and boosting efficiency with automated monitoring
Oligonucleotide synthesis is almost exclusively conducted using automated solid-phase methods. This involves resin beads mounted on a machine being treated in sequence with various reagents and washes to build up the oligonucleotide from its constituent nucleotides. Wash solvents, solutions of reagents, as well as the nucleotide bases themselves, require manual preparation at specific concentrations and placing on the equipment such that the control software can correctly identify each reagent and use it in its assigned place.
The requirement for manual human input in this process is both onerous and a potential source of error. To alleviate this, computer vision can be applied in the laboratory by placing small cameras in convenient locations such as the ceiling, walls and on the scientists themselves, where they can observe the movements and actions in their field of view, alongside the automated processes carried out by laboratory equipment. An example from this stage would be identifying materials by reading the names, CAS numbers, lot numbers, expiry dates and other relevant data located on labels when being handled by the scientist – all of which can all be noted and linked back to inventories and experimental records. By watching the bench where the preparations of these reagents are carried out, validation of volumes and types of solvents and reagents used can be achieved and cross referenced against the known expected values. If deviations from these are detected, this can be flagged to the scientist to check the process using the footage itself to pinpoint errors.
In addition to interpreting labels and manipulations on a single bench, materials can also be followed on their journey around the laboratory from bench to machines, and back again. This involves placing further wide-angle cameras to enable full coverage of the lab, and utilising state of the art object detection models (trained on proprietary data) and tracking algorithms. Data from this would be valuable for gaining greater understanding of processes, increasing efficiency, and ensuring compliance with procedures and traceability at all stages in the process.
Colour change tracking
Once the reagents have been prepared, the actual oligonucleotide synthesis can occur, and this is conducted using a specialised machine. The process consists of treating a solid resin covered in oligonucleotide attachment points with various solutions in a specified sequence to build up the desired oligonucleotide base by base. Firstly, trichloroacetic acid is added to remove the dimethoxytrityl (DMT) protecting group end of the growing oligonucleotide and expose the reactive hydroxyl (Figure 1, stage 1).
A nucleotide base is then added in a suitably reactive (phosphoramidite) form, which bonds to the hydroxyl to form the next base in the chain (Figure 1, stage 2), followed by an oxidation to form the phosphate on the backbone (Figure 1, stage 3). This process is repeated multiple times with the order of nucleotides applied corresponding to the base sequence of the desired oligonucleotide. It happens that the protecting group in the modern phosphoramidite process is almost always DMT, which, when removed, forms a very highly coloured orange species. Given this deprotection step is a prerequisite for a further nucleotide base being successfully added to the oligonucleotide chain, the appearance of the orange colour can be treated as a validation that the desired deprotection has happened. The colour change can be easily detected via computer vision based dominant colour analysis to determine whether the correct solvents and reagents have been applied to the resin, and that the chain is growing as anticipated. If the colour change is not detected, then recording, pinpointing and alerting the user where the synthesis has failed would ease the burden of troubleshooting considerably.
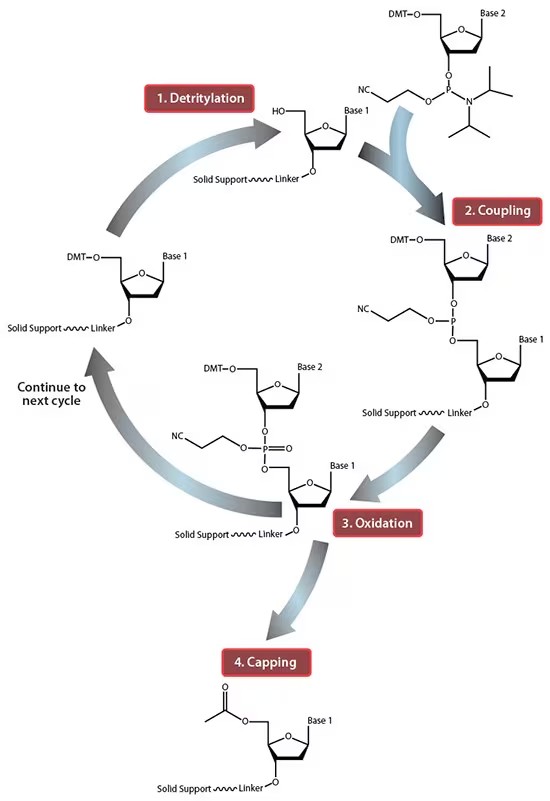
Figure 1: Simplified phosphoramidite method2

Figure 2: An example of a computer vision system able to identify and track hands, pipettes, vials and the liquids within them in real-time
Object tracking and cross-referencing
Once the synthesis is complete, the resin with the synthesised oligonucleotide chains attached is removed from the plastic column and suspended and heated in a solution designed to cleave the oligonucleotide from the resin support. This process also removes the remaining protecting groups to leave the free oligonucleotide in solution. Depending on the specifics, this reaction may also result in a colour change which can be identified and characterised as outlined above. When the cleavage is complete, the free oligonucleotide in solution is filtered away from the now empty resin beads. This cleavage and deprotection process therefore involves transferring an oligonucleotide between three separate containers and, while relatively trivial for a single synthesis, in practice synthesis are usually performed in parallel. For example, if using a synthesiser with 12 positions, 36 separate containers would result by this stage, each requiring diligent labelling and tracking of the current contents and of which material went into which container to ensure that the actual contents of the final container are correctly identified.
The ability to track individual objects and cross reference their labels could therefore be transformational in this type of environment. Eliminating uncertainty as to the materials being used at any point greatly reduces the chance of mistakes in identification of reagents or samples. By applying this to individual oligonucleotide sample containers, at all stages the identity of each sample being used can be logged, not only tracking its location through the lab but also its current observable state and the container it resides in.
This would be a major boon in busy labs as multiple samples being moved around simultaneously can appear to a human to be identical, making them near impossible to distinguish without accurate labelling. By following the sample from its first step until it is placed in its final container, it would be possible to create a log of events pertaining both to its identity and exactly what operations were performed to produce it, ensuring full traceability (Figure 2).
Progress tracking
The next task involves analysing the free oligonucleotide, firstly by UV-Vis spectrometry to confirm whether DNA or RNA is present in the solution, and then by mass spectroscopy to confirm the largest molecular mass present conforms with the expected oligonucleotide fragment, and quantify the presence of smaller fragments and impurities. Lyophilisation to remove any solvent may also be performed at this stage. Once information has been gathered on the identity and purity of a particular oligonucleotide, the last step is to purify it by preparative HPLC and balance the salt content, often using ion exchange processes. The purification process outputs a series of fractions with varying content, again multiplying the number of containers and the complexity of managing them. These samples and fractions can be identified and tracked around the lab as outlined above, but also potentially linked to the output of the analytical equipment, ensuring that data associated with all materials can be stored ready for easy examination when required.
In addition to the use of computer vision techniques to streamline a process such as oligonucleotide synthesis, non-vision AI tools can also be used to keep track of the progress of an experiment. By providing a system utilising large language model (LLM) agents with an explanation of the process to be carried out in advance, it is able to break down the process into discrete nested actions, steps and stages. These individual actions can then either be checked off manually by the scientist, or automatically as the computer vision model recognises them being performed. This allows for the creation of an audit trail of events explaining exactly what happened, when and where, and how it compares with the planned protocol. This trail comes complete with videographic evidence, providing yet another opportunity to reduce human error, record deviations from a known method, and, in the event of an unexpected result, allow in-depth investigation of what actually happened. Specific examples in oligonucleotide synthesis include detecting whether deprotection steps have completed successfully, and whether the correct nucleotide bases have been added in the correct sequence, whether the label on a sample accurately reflects the contents, and whether analytical data is associated with the correct sample. In identifying failures during synthesis and purification, it would be possible to stop failed or mislabelled syntheses to prevent the waste of potentially expensive reagents, and save time working up. Perhaps even more important is the vastly greater information available from a single failed (or successful) experiment when conducted using computer vision – and the ability to use this information to find the source of the failure or success and learn from it – rather than conducting lengthy practical investigations into an issue with no guarantee of success.
Summary
Computer vision has huge potential for simplifying, facilitating and generally relieving some of the heavier burdens from scientists working in oligonucleotides. Perhaps the most transformational aspect of computer vision is in the extra data, understanding and insights it can capture during lab work which would otherwise go unnoticed or unnoted, and the consequent ability to produce better conclusions based on a richer understanding of the processes under study.

Cei Provis-Evans, associate principal scientist at Reach Industries, brings 16 years of diverse experience across the chemical industry and academia. With a PhD in Organometallic Chemistry from the University of Bath, UK, Cei’s earlier career included pivotal roles in analytical chemistry and formulation science at GSK and the British Pharmacopoeia Lab. His journey has since spanned analytical and process chemistry at leading pharmaceutical CROs, Pharmaron and CatSci Ltd. Together with the team at Reach Industries, Cei is on a mission to supercharge modern science through innovation and expertise.

Sam Whitmarsh, director of Analytical Science & Digital Transformation at CatSci, and founder and science lead at LabLinks, has 20 years of R&D experience at AstraZeneca, BP and CatSci. After completing his PhD at the University of Bristol, UK, he held roles in analytical science at AstraZeneca, founded BP’s UK mass spectrometry facility, and led the BP Global Analytical Science Network. Sam joined CatSci as head of scientific operations in 2020 and became director of Analytical Science and Digital Transformation in 2022. Sam is a Royal Society of Chemistry fellow serving with the Separation Science (SSG) and Chemical Information and Computer Applications (CICAG) interest groups.