MALDI Imaging and Spacial Protemoics
Technological advances in MALDI imaging and spatial proteomics for medical research
Recent advancements in MALDI mass spectrometry imaging are transforming disease understanding and treatment. This innovation promises rapid diagnostics and personalised medicine, with reliable imaging within five years

Andreas Sonnen at University Medical Center Utrecht, Michael L Easterling at Bruker Daltonics
Recent technological advances have improved the ability of medical researchers to deepen our scientific understanding of the causes of disease and possible treatments, while continuing to support efforts to improve patient care.
One such innovation is matrix-assisted laser desorption/ ionisation (MALDI) mass spectrometry (MS) imaging, which has shown potential for improving diagnostic speed and accuracy in medical applications.
Technological innovations in this technique provide a molecular profile of thousands of molecules at each image pixel without the loss of tissue morphology at increasing acquisition speeds and spatial resolutions. This opens the way to assess, for example, molecular tumour heterogeneity together with classical histology on the same image by carefully selecting peaks from the measured mass spectra.
MALDI MS imaging enables visualisation of the spatial distribution of proteins, peptides, lipids and small molecules within thin slices of tissue. When looking at the proteome, lipidome or metabolome, the spatial distribution of compounds contains valuable information.
Spatial omics applications have significantly expanded insights into the causes of disease, as well as improving understanding and therapeutic treatment evaluation for a wide range of disorders, particularly cancer.
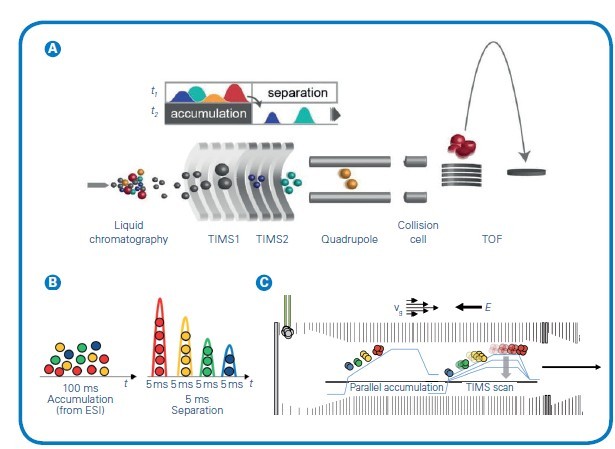
Figure 1: (A) The timsTOF platform's dual-TIMS analysers allows for parallel accumulation followed by high-resolution ion mobility separation. The ion cloud enters TIMS1 over a 20-100ms accumulation time but elute from TIMS2 as discrete ion packets separated by CCS and compacted into 2-5 ms segments before entering the TOF. (B) This temporal preconcentrating effect of dual-TIMS funnel results in up to a 30x signal-to-noise improvement compared with other continuous acquisition instruments. (C) Simultaneous filling of TIMS1 while eluting from TIMS2 delivers 100% duty cycle. Adapted from Meier et al, bioRxiv (2020)
Unravelling disease mechanisms to develop novel therapeutics
MALDI MS imaging has emerged as an important, rapid and accurate tool for medical research. Researchers are studying how a patient’s gene variations may guide the selection of drugs or treatment protocols that minimise harmful side effects or ensure more successful outcomes.
Additionally, such personalised medicine approaches could also indicate an individual’s susceptibility to certain diseases, allowing physicians to monitor and possibly prevent the manifestation of a future health condition.
Current research focuses on implementing MALDI imaging and MS for better clinical diagnostics within a pathology department, which has complex needs, standard operating procedures and specific regulations to follow.
The long-term goal is to not only use these tools to study various diseases, but also to establish a reliable diagnostic imaging modality within the next five years.
The microenvironment of tumours consists of a combination of connective tissue, blood vessels and inflammatory cells in different ratios. Each of these components has its own unique molecular signature.
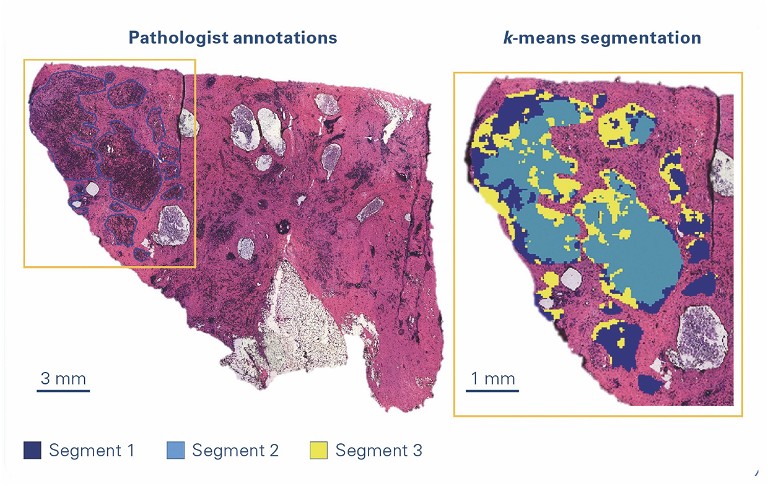
Researchers often rely heavily on interpreting tissue pathology and creating molecular maps to bridge the gap between ‘omics’ and traditional histology. It is now possible to find specific molecular biomarkers for different subtypes of tumours of various prognostic subclasses.
There are ongoing MALDI MS imaging studies to further subclassify and determine biomarkers for these subclasses.
Technological innovations
Rapid technological advances have enabled progressively sophisticated and robust genomic, transcriptomic, proteomic and other ‘omics’ analyses. However, to gain a deeper and more comprehensive understanding of biological systems, the spatial context of the resulting data and information has become increasingly important.
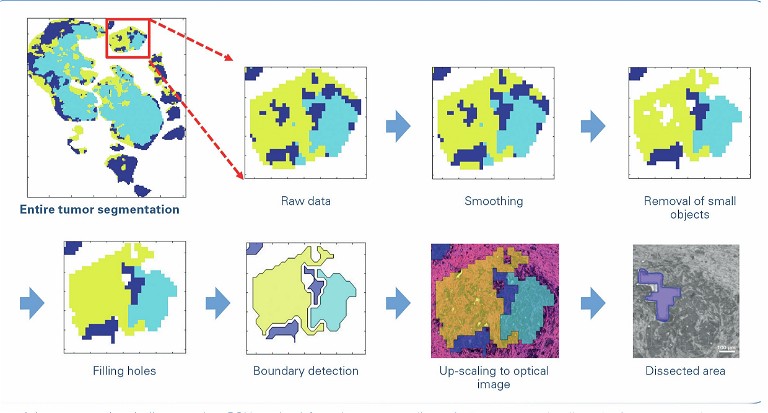
Researchers have trialled multiple solutions for performing various types of spatial – omics analyses – including spatial genomics and spatial proteomics. Among other methods, the combination of laser microdissection with trapped ion mobility spectrometry (TIMS) enhanced MALDI MS imaging, which provides a collisional cross-section (CCS) to enhance specificity while, in parallel, improving sensitivity via ion accumulation.
Laser-capture microdissection
Laser-capture microdissection (LCM) is a minimally disruptive method to obtain cytologically and/or phenotypically defined cells or groups of cells from heterogeneous tissues. It is a versatile technology that allows the preparation of homogenous isolates of specific subpopulations of cells from which RNA/DNA or proteins can be extracted from a wide variety of tissue samples. The sample is then available for further methods such as proteomics and other analytical techniques. Laser microdissection is now used in many research fields, eg, neurology, cancer research, plant analysis, forensics and climate research.
TIMS
Ion mobility spectrometry (IMS) is a powerful analytical technique that has been widely applied over the last five decades, primarily in chemical physics and analytical chemistry applications. Only recently has IMS technology improved to the point where it can be used to enhance separation, identification, and quantification of peptides and proteins in modern workflows.
TIMS is an IMS separation technique in the gas phase, which resolves sample complexity with an added dimension of separation in addition to high performance liquid chromatography (HPLC) and MS, increasing peak capacity and confidence in compound characterisation. TIMS also serves to accumulate and concentrate ions of a given mass and mobility, enabling a unique increase in sensitivity and speed along with the additional dimension of separation. Ions are propelled through the TIMS tunnel by a gas flow. While the ions are accelerated by the gas, a counteracting electrical field gradient is trapping the ions in the gas stream, separated based on their size-to-charge ratio described by the mobility (1/K0). Ramping down the electrical field allows for selectively releasing ions from the TIMS tunnel according to their mobility (see Figure 1).
Understanding complex samples requires the analytical depth provided by the ion mobility option of TIMS, in addition to mass and charge. The analyses of samples such as tissue sections benefit from TIMS because it can separate isobaric or isomeric metabolites, lipids, peptides or glycans to get the true spatial localisation of an analyte. This technology offers a chance to differentiate isomeric distributions where high mass resolution fails. MALDI workflows can take advantage of the most advanced features found in TIMS based on the CCS of detected molecules. TIMS allows for the separation of molecules depending on the shape of the ions. Ions enter the dual TIMS device together with a gas stream and are accumulated in the first section by an electrical field.
The actual separation takes place in the second part of the TIMS cartridge, where ions are eluted in a temporal and spatial fashion by lowering the electrical potential. Variable ramp speed and mobility range adaptation give rise to the optimisation of different classes of molecules, allowing high flexibility for the user.Using CCS as an outcome of TIMS to validate the identity of an analyte offers researchers an additional quality criterion. CCS-aware software intelligently matches spatial MALDI-TIMS imaging data with -omics results and enables vital morphological context to identification lists.
Another important area that would be impacted by the potential diagnostic value of omics imaging is discrimination between early (T1-) colorectal carcinomas that do or don’t require invasive surgery for lymph node resection. By combining MALDI MS imaging and TIMS, researchers hope to identify determinants of metastatic spread within the tumour and in its microenvironment. Currently, the vast majority of lymph node resections turn out not to contain any metastasis, so identification of such determinants will lead to an enormous reduction in surgical procedures and associated morbidity and costs.
MALDI MS
MALDI MS analyses have been used for more than 25 years. With inherent benefits including its speed of analysis and application flexibility, the convenience and effectiveness of MALDI MS is magnified by virtue of its low sample volume requirements and broad sample content (salts, buffers) amenability.
Dramatic increases in the depth of MALDI analyses have been made routine by intuitive software, and easy automation allows for high throughput solutions. The sample for analysis by MALDI MS is prepared by mixing or coating with a solution of an energy-absorbent matrix, which entraps and co-crystallises the sample when dried.
The matrix is ionised with a laser beam, and transfers the charge to the analytes, generating singularly charged ions from analytes in the sample that are then accelerated into the mass spectrometer. Ions are separated from each other on the basis of their mass-to-charge ratio (m/z) before being detected and measured using the mass analyser. The high throughput and speed associated with MALDI MS have made this technology an important tool for large-scale proteomics, imaging studies and other analyses.
“ Understanding complex samples requires the analytical depth provided by the ion mobility option of TIMS, in addition to mass and charge ”
Expanded capabilities
In one such study that focuses on cancer treatment, LCM can be used to extract tissue for further analysis by MALDI MS to get spectral information, to create a molecular map of the tumour and the tumour heterogeneity. Based on such a molecular map, physicians in the future could decide which treatments are suited for the patient.
Because cancer cells have significant genetic and epigenetic modifications that influence protein expression and their metabolic state, it’s possible that finding new targets or new molecular markers for these tumours can help researchers develop other treatment possibilities in the future.
Researchers are working towards visualising lipid and protein modifications generated by plasma treatment, mostly oxidation. The combination of MALDI imaging and TIMS has helped move this study further with the ability to spatially resolve the oxidation products, which was not possible before.
While much progress has been made, these technological advances continue to push boundaries by unravelling the molecular mechanisms that underpin diseases and by finding new approaches for treatments that will improve patient outcomes. The long-term goal is to take these medical discoveries from the laboratory and eventually move them into clinical practice.
Together, they hold potential in the advancement of personalised medicine, which experts believe could result in new therapies that can improve a patient’s response and ensure better care.
When looking at medicine in a personalised care setting, leveraging the full potential of all available data is of the utmost importance to select the proper treatment for each patient and prevent unwanted treatments, thus saving overtreatment for the patient and costs for society.
“ The long-term goal is to take these medical discoveries from the laboratory and eventually move them into clinical practice ”
Researchers see MALDI MS imaging as a tool for personalised medicine and not simply as a potential substitute for immunohistochemistry or other techniques. The result will be a new way to provide healthcare, where physicians can classify and treat diseases based on their molecular profiles, instead of simple physical indications and symptoms.
Combining this knowledge with a patient’s genomic information may help medical professionals make more effective decisions for the individual patient. By enabling each patient to receive earlier diagnoses, risk assessments and optimal treatments, personalised medicine holds promise for improving healthcare while also lowering costs. Additionally, as the treatments become more specialised, more information about the patient will become key to the successful application in a clinical setting.
References
- Feucherolles M, Frache G (2022) 'MALDI Mass Spectrometry Imaging: A Potential Game-Changer in a Modern Microbiology,' Cells, 2:11 (23), 3900
- Flach RN et al Fransen NL, Sonnen AFP, Nguyen TQ, Breimer GE, Veta M, Stathonikos N, van Dooijeweert C, van Diest PJ' (2022) 'Implementation of Artificial Intelligence in Diagnostic Practice as a Next Step after Going Digital: The UMC Utrecht Perspective. Diagnostics' 21:12(5), 1042, Basel.
- Meier F Brunner A-D et al Frank M, Ha A, Bludau I, Voytik E, Kaspar-Schoenefeld S, Lubeck M, Raether O, Aebersold R, Collins BC, Röst HL, Mann M (2020) 'Parallel Accumulation – Serial Fragmentation Combined with Data-Independent Acquisition (DiaPASEF): Bottom-up Proteomics with near Optimal Ion Usage' (2020), bioRxiv, 656207. https://doi.org/10/ggr98g

Dr Andreas Sonnen MD is an assistant professor in the Department of Pathology at the University Medical Center in Utrecht, the Netherlands. He studied of biochemistry and chemistry at Constructor University in Bremen, Germany, before completing his DPhil. in Biochemistry at the University of Oxford, UK, in 2009. He worked as a Sir Henry Wellcome post-doctoral fellow at the Max Planck Institute in Munich, Germany and Division of Structural Biology in Oxford, UK. He ended his fellowship early to study medicine at the University of Freiburg and the University of Heidelberg both Germany, while working as a post doc at the Center for Chronic Immunodeficiency (Freiburg) and later EMBL in Heidelberg. After moving to Utrecht, he first worked in the group of Prof Friedrich Förster before taking up his role as assistant professor and MD in the department of pathology.

Dr Michael Easterling is vice president of imaging at Bruker Daltonics. He received his PhD from the University of Georgia, United States in 1998. After moving to IonSpec, in 1998, Dr Easterling was responsible for commercial demonstration capabilities and played a role in instrument and applications development creating relevant proof statements for ICR technology. In 2001, Dr Easterling moved to Bruker Daltonics to head the applications division for the MRMS product line where he is also responsible for maintaining collaborative efforts with investigators in both academia and industry. In 2016 the MRMS business unit merged with the MALDI imaging programme for which Dr Easterling served as director.