Analytics: Mass Spectrometry
Metabolomics and Lipidomics Research Accelerated by Trapped Ion Mass Spectrometry
Innovations in cellular biochemistry are helping to bring better personalised medicine onto the market. How can trapped ion mass spectrometry (TIMS) support this moving forward?
Lucy Woods at Bruker Daltonics
Metabolites hold the key to understanding cellular biochemistry. Metabolomics research is, therefore, crucial for obtaining fundamental insights into disease pathogenesis and discovering biomarkers for disease diagnosis. As the product of metabolic pathways, metabolites include small molecules such as lipids, sugars, nucleotides, and amino acids. Advances in other ‘omics’ fields, such as genomics and proteomics, have paved the way for metabolomics and, more recently, lipidomics, to play a major role in clinical research and practice, with future potential in personalised medicine.
‘Global’ or untargeted metabolite profiling is revealing new discoveries that link cellular pathways to biological mechanisms, shaping the understanding of cell biology, physiology, and medicine. However, metabolic pathways involve isomer metabolites – structural variations of the same metabolite – which complicates their characterisation. The presence of isomers makes reliable compound identification and accurate interpretation of data very difficult.
Mass spectrometry (MS) is a mainstay in metabolomics, due to its ability to quantitatively measure thousands of metabolites from small amounts of biological material. But, even high-performance mass spectrometers struggle to analyse separate isomers due to the chemical similarity of isomer metabolites. Because isomers can have the same mass, charge, and physical properties, separation and identification are time- and resource-intensive processes.
In addition, the metabolome is extremely sensitive; a person’s metabolic phenotype is influenced by environmental factors and holds enormous variation. Data can also be more ambiguous because metabolites are the products of upstream biological transformations of molecules in a similar mass range, which requires higher sensitivity to obtain broad metabolite coverage and relative concentrations.
Overcoming the Isomer Challenge
Ion mobility spectrometry (IMS) has long been used for the post-ionisation separation and structural characterisation of biomolecules, and its combination with MS has provided researchers with the tools to separate molecules in a third dimension, using collisional cross sections (CCS). IMS-MS not only allows for the measurement of ion size and the separation of isomers with the same mass-to-charge (m/z) ratio, but increases dynamic range and peak capacity compared with MS alone.
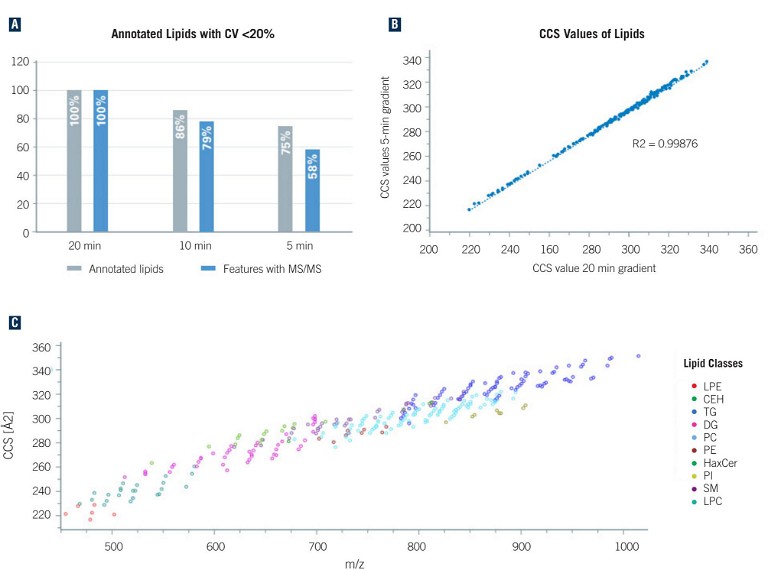
Figure 1:
A) Overview of the number of annotated unique lipids at different LC runtimes from merged data of both polarities. Reducing the LC runtime by a factor of 4, the number of annotated lipids decreases to 75%. The relative values refer to 20 min (362 lipids, 7592 features with MS/MS)
B) Comparison of observed lipid CCS values from 5- and 20-minute gradient run. The plot demonstrates the power of CCS values as a qualifier for compound identification as the CCS values are independent of the chromatographic setup
C) Distribution of the detected lipids in the m/z and CCS space (5-minute runs, ESI-[+]). For different lipid classes, clear trends can be observed that enable validation of the annotations
It has, therefore, emerged as a powerful technology to address the isomer metabolite challenge, and the use of IMS-MS in untargeted metabolomics now supports metabolite identification through the generation of orthogonal data (CCS values).
New technology is continually being developed to achieve a deeper understanding of the biological relevance of metabolites in health and disease. Since its commercialisation in 2016, trapped ion mobility spectrometry-MS has contributed to significant advances in biomolecule separation and characterisation. Its higher mobility resolving power enables the separation of molecules with differences in their CCS values so small that traditional IMS-MS cannot distinguish them. In addition, the novel scan mode, termed ‘parallel accumulation – serial fragmentation’ (PASEF), multiplies the sequencing speed without any loss in sensitivity (1).
TIMS in Lipidomics
The acceleration of metabolomics research has driven new developments in lipid biochemistry. Lipids are fatty small molecules that share common physical and chemical properties, and the abundance of lipids and different lipid classes are key to metabolic regulation. As metabolomic possibilities progress, lipidomics becomes a prominent source of collecting and measuring data within the metabolome. Due to lipids’ high isomeric content, and their varying abundance in typical samples, such as plasma extracts, analytical equipment with high sensitivity and dynamic range is required.
“ Scientists are looking further into metabolomics to examine how the cells in our body behave and what this could mean as we look towards a future of personalised medicine ”
Recently, a 4D-Lipidomics workflow has been demonstrated to simplify the annotation and validation process of lipid isomers using TIMS powered by PASEF (2). In this study, the lipid content of a NIST SRM 1950 extract from reference plasma (Sigma-Aldrich, Germany) was profiled using the 4D-Lipidomics workflow, which was comprised of: reverse phase liquid chromatography (RP-LC) (Elute UHPLC system, Bruker, Germany); TIMS-MS and MS/MS data acquisition in PASEF MS/MS mode (timsTOF Pro, Bruker, Germany); and 4D data processing or m/z, mobility, and intensity using MetaboScape 2021b software (Bruker, Germany).
To investigate the increased MS/MS quality and peak capacity provided by PASEF, the SRM 1950 lipid extract was analysed with different chromatographic run times of five, ten, and twenty minutes.
The maximum number of unique identified lipids (362) was observed with 20-min gradient times and, even when reducing the runtime to 5 mins, 75% (271) of the annotated lipids with a CV of less than 20% were still detected (see Figure 1A). A comparison of the measured CCS values from the 20-min and 5-min gradients depicts a strong correlation (see Figure 1B), which demonstrates that, in contrast with retention times, CCS values are independent of LC, and can be reliably used for compound identification (see Figure 1C).
The results highlight the high MS/MS coverage that can be achieved using PASEF, and how automatically acquired CCS values increase confidence in annotations and compound identification. These results set the stage for deep lipidomics profiling, through methods like 4D-Lipidomics at high throughput, to enable studies that require a high turnover – for example, clinical research studies.
Looking Towards the Future of Personalised Medicine
Molecular alterations at the metabolome level reflect disturbances in preceding biological cascades, bridging the gap between the genome and phenotype. Changes at this level can lead to the onset of disease symptoms, making metabolomics an essential diagnostic and prognostic tool in investigating the mode of action of chemical compounds and obtaining an in-depth understanding of the impact of infection, for example (3). Current metabolomics research focuses on a range of challenging diseases, including Alzheimer’s disease and cancer, and scientists are looking further into metabolomics to examine how the cells in our body behave, and what this could mean as we look towards a future of personalised medicine.
As new technologies evolve and researchers look to optimise equipment to accelerate their studies, it is important that software and analytics keep pace to support the abundance of new data. Advances in MS technology such as TIMS bring researchers one step closer to a future in which personalised medicine is within reach.
References
1. Meier F et al, Online Parallel Accumulation—Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer, Technological Innovation Resources, 2018; 17(12): pp2,534-2,545.
2. Zubeil F et al, 4D-Lipidomics™ workflow for increased throughput, Bruker Daltonics, App note LCMS-179, 2021.
3. Tounta V et al, Metabolomics in infectious diseases and drug discovery, Molecular Omics, 17(3): pp376-393, 2021.

Lucy Woods, PhD, is Business Unit Manager Phenomics and Metabolomics at Bruker Daltonics. She has been working for Bruker Daltonics for six years, prior to which she worked for pharma companies GlaxoSmithKline & AstraZeneca developing workflows to identify unknown impurities using mass spectrometry.
Lucy has a strong background in ion mobility-mass spectrometry, having completed her PhD under the supervision of last years’ Thomson medal winner, Professor Alison Ashcroft at the University of Leeds, UK. This led Lucy to join Bruker and take the role of Product Manager for the timsTOF series, where she was responsible for launching the timsTOF Pro and timsTOF fleX platforms.
Lucy is now leading the Global Business Unit focussing on Metabolomics, where she is responsible for the development and execution of new workflows for metabolomics and lipidomics using the timsTOF platforms.